LIDC XML Simple Manual
NEW, CODE added, compatible with MITK_Version >= 2016.05
Firstly, compile and build the MITK according to manuals on mitk.org, make sure the lib.org.lidcxml is compiled.
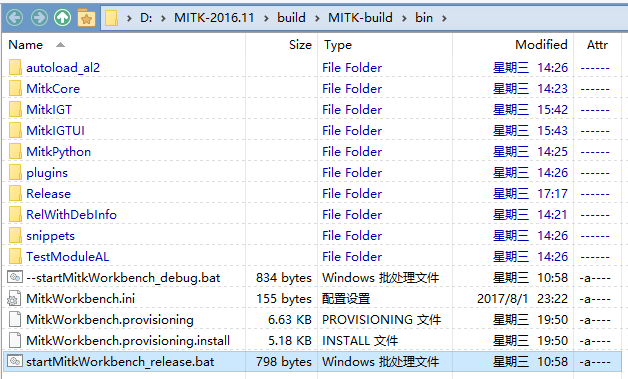
Start the startMitkWorkbench_release.bat in buid/mitk_build/bin
Load the original LIDC datasets
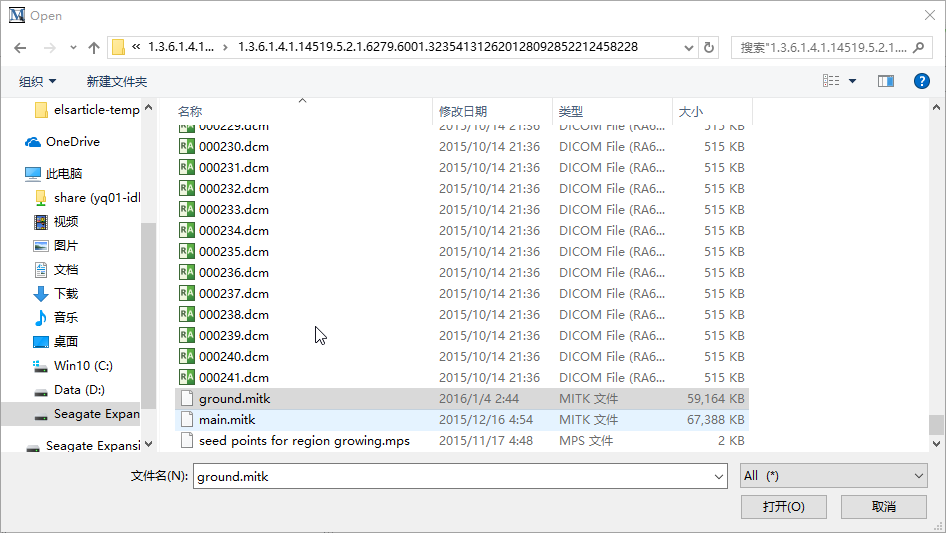
select one of the .dcm file to open them in mitk
You can get this page (right click-open in new tab to see the original image)
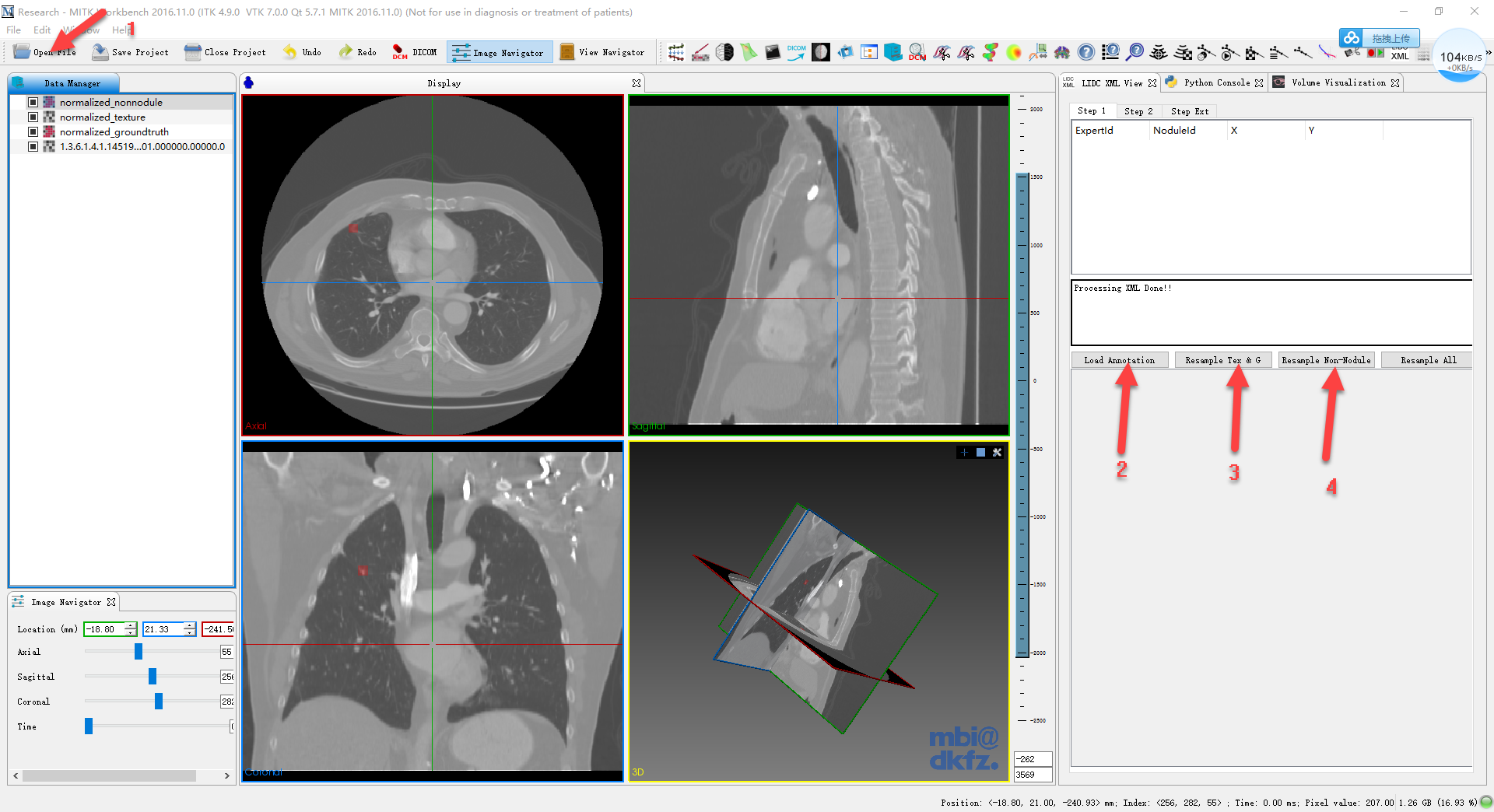
the 1st arrow, open the data set
after opened the data, select it in data manager, and click 2nd arrow, load annotations
then, the annotation (.xml in corresponding lidc data sets) will be loaded
since original data sets are not isotrophic, we need resample them first. use Resample Tex&G then Resample Non-nondule or just click on Resample All which actually consists these two steps.
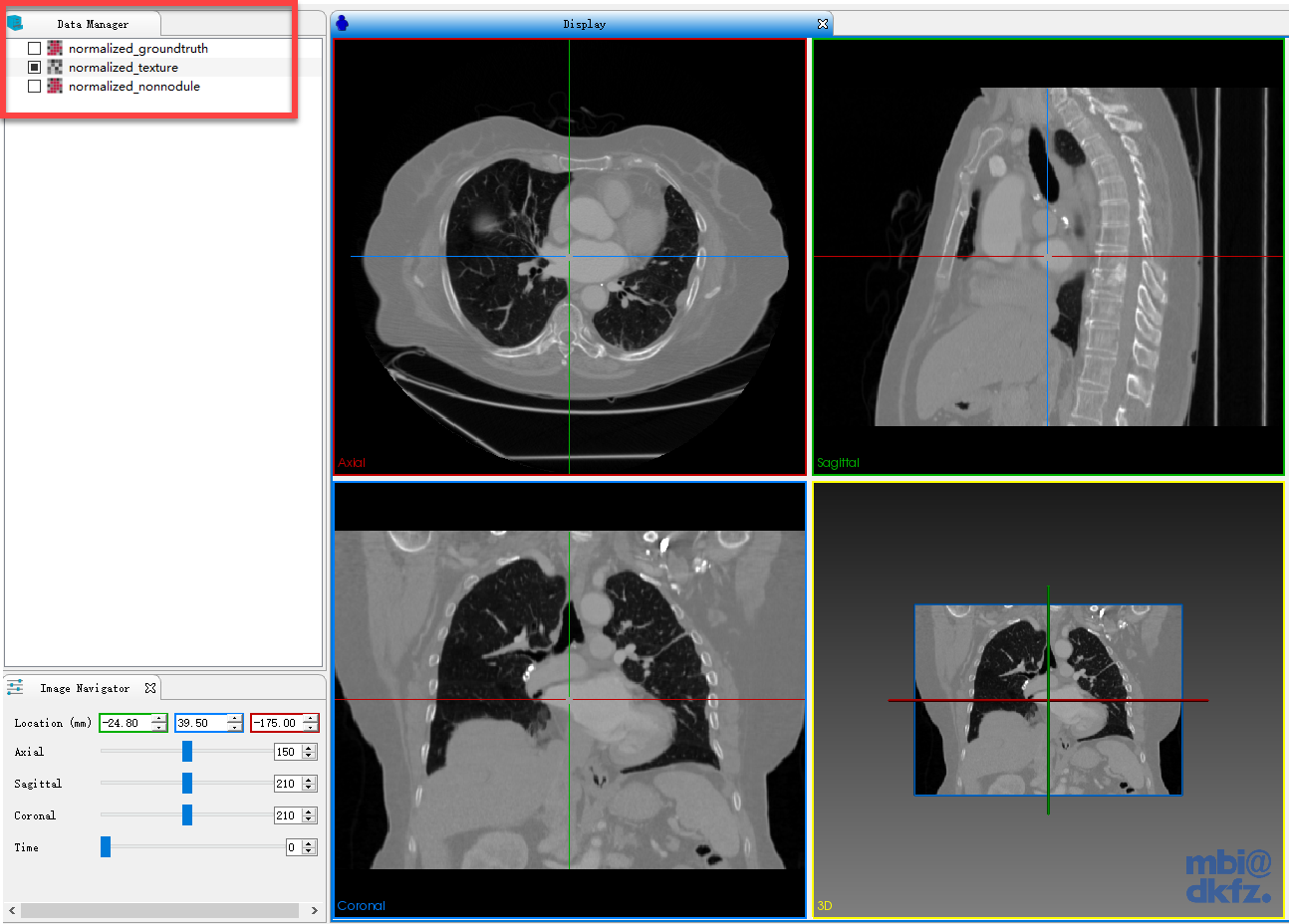
wait one minute and you get three new data in data manager, normalized_texture, normalized_groundtruth and normalized_nonnodule
then, click on step2 on the right tab.
here you go
make the original data set (here is the 1.5.1.4.xxxxxxx) selected and press the Select Working Dir and select the corresponding image in the dropboxes (texture--->texture, ground--->ground, nonnodule--->nonnodule), (right click-open in new tab to see the original image)
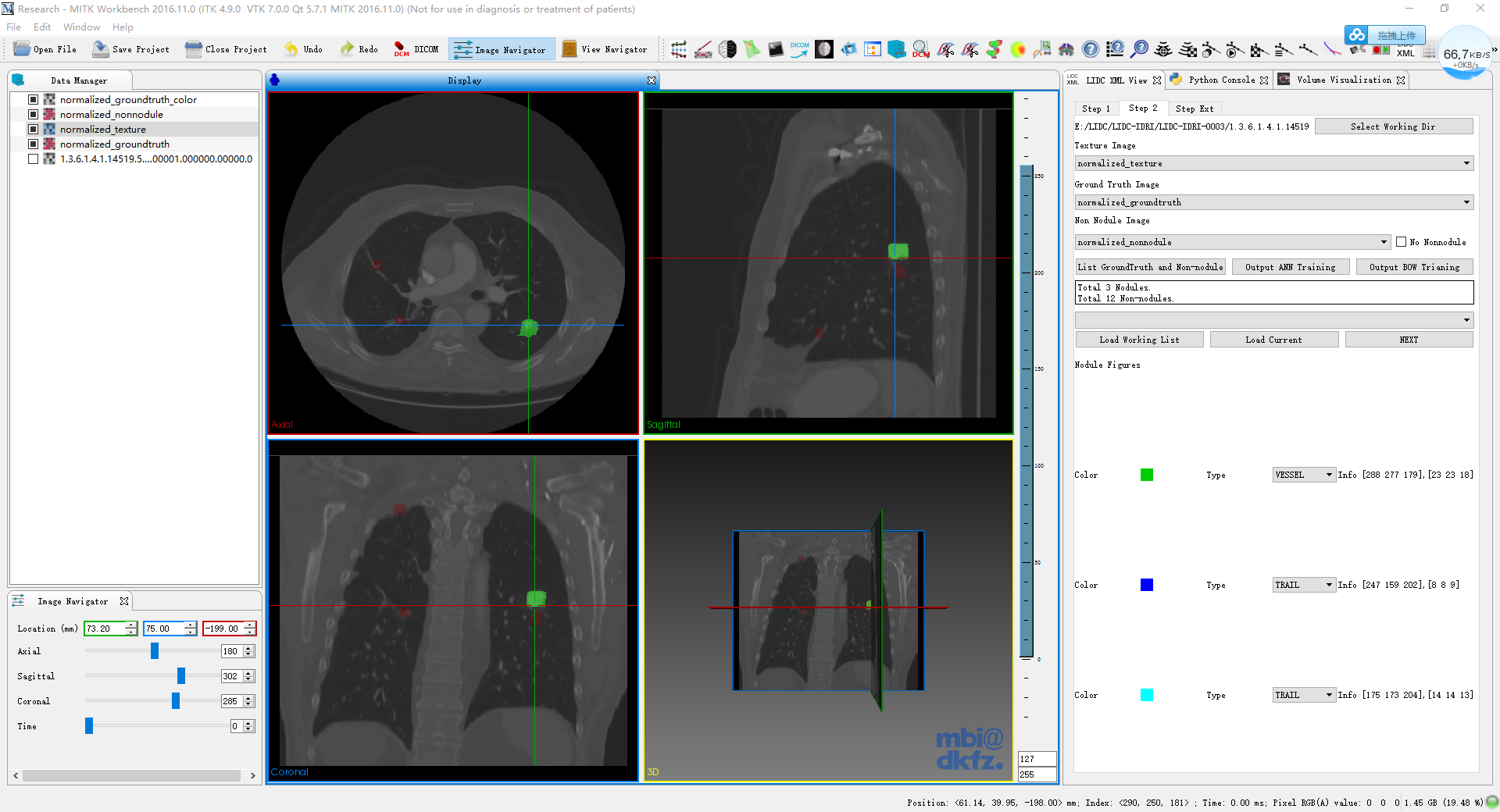
finally, click on the List Groundtruth and Non-nodules, the corresponding nodules will be shown.
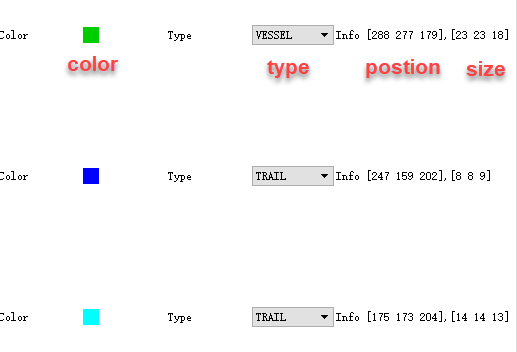
- in the list, the color of corresponding nodule is shown and followed by the nodule type. the infor shown the position and size of the nodule.
- for my use, i have written the output ANN training and output BOW training two functions corresponding to the two buttons on the page, for more uses please change the code.
I suggest to resample all original images as a united .mitk file including the texture-groundtruth-nonnodule.